LoRA, A Long Read Assembly pipeline for microbiome data
What is Nephele?
Nephele is NIAID’s web application for microbial genomics and microbiome analysis which can be found at nephele.niaid.nih.gov.
We have pipelines for QC of sequenced reads, amplicon metagenomics, shotgun metagenomics, as well as SARS-CoV-2 viral variant analysis.
What is LoRA?
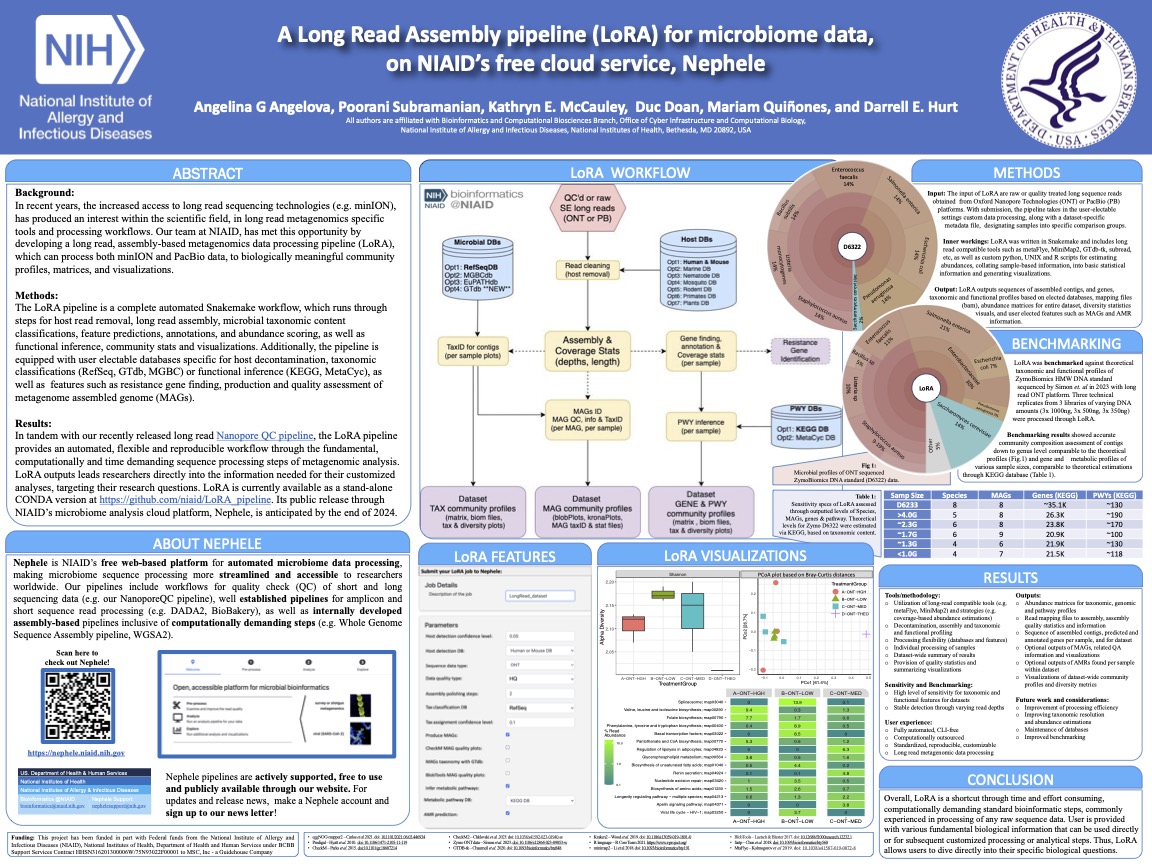
The LoRA pipeline is a complete automated Snakemake-based workflow for long read metagenomic sequencing data processing. It runs through processing steps of host sequence read removal, long read assembly, microbial taxonomic content classifications, feature predictions, annotations, and abundance scoring, as well as functional inference, community stats and dataset visualizations. The pipeline is equipped with user electable databases specific for host decontamination, taxonomic classifications (RefSeq, GTdb, MGBC) or functional inference (KEGG, MetaCyc), as well as features such as resistance gene finding, production and quality assessment of metagenome assembled genome (MAGs).
The LoRA pipeline will soon be available publicly and freely through NIAID’s microbial analysis platform Nephele. Meanwhile, an HPC clusterized version of the pipeline is available at this GitHub repo for LoRA
Visit our poster during ASM NGS 2024,
at the Hyatt Regency Washington on Capitol Hill in Washington DC (400 New Jersey Ave NW, Washington, D.C. 20001),
on Monday Oct 14th,
In poster session I, between 2:45-5:15pm
At Poster Board number: PIP-MON-135.